Working with Images
In this tutorial we’ll provide an overview of the concepts and classes that represent medical images in the imfusion-sdk and how to work with them.
[ ]:
import _engine_plugin_guard
import imfusion
import imfusion.machinelearning as ml
import numpy as np
from matplotlib import pyplot as plt
from ipywidgets import IntSlider, FloatSlider, interact, fixed
from demo_utils import unzip_folder, mpr_plot
unzipped_folder = unzip_folder('data/pet-ct-rtstruct.zip')
Image classes
SharedImage: class holding a single image, can be a 2d Image (X-Ray, Ultrasound) or a 3d (CT, NM, MR, …) volume. The word “Shared” refers to the fact that it can have multiple representation on different devices (typically CPU + GPU via OpenGL) which are kept automatically in sync.SharedImageSet: A Set of SharedImage. This is the main entry point of any method in the ImFusion SDK which operates on image data. For volumetric data, a SharedImageSet mostly comprises a single SharedImage representing a volume (i.e. a CT), but it is fundamental for holding image series consisting of multiple frame, which have no trivial 3d representation, i.e. an ultrasound acquisition, an OCT pullback, a Fluoro navigation, a 4D-CT and so on..ImageDescriptor: class describing the basic properties of the image, like its storage type (int, float, …), its spacing, its shape.Matrix: orientation of the image data in the physical world
Loading Images
Since ImFusion files (.imf) can hold several SharedImageSets, loading an
.imffile returns a list ofSharedImageSetsA
SharedImageSetbehaves like a list. You can iterate over it or index individual elements
[2]:
pet, *_ = imfusion.load('data/pet-ct-rtstruct/pet')
labels, *_ = imfusion.load('data/pet-ct-rtstruct/tumors.imf')
print('PET: ', pet)
print('Labels: ', labels)
# The loaded SharedImageSets each contain one images.
print(f'Num of images in PET: {len(pet)}, num of images in Labels: {len(labels)}')
# Get SharedImage from SharedImageSet
image = pet[0]
print('SharedImage:', image)
PET: imfusion.SharedImageSet(size: 1, [imfusion.SharedImage(FLOAT width: 192 height: 192 slices: 227 spacing: 3.64583x3.64583x3.27 mm)])
Labels: imfusion.SharedImageSet(size: 1, [imfusion.SharedImage(UBYTE width: 192 height: 192 slices: 227 spacing: 3.64583x3.64583x3.27 mm)])
Num of images in PET: 1, num of images in Labels: 1
SharedImage: imfusion.SharedImage(FLOAT width: 192 height: 192 slices: 227 spacing: 3.64583x3.64583x3.27 mm)
[DICOM] Detected different rescale values per frame, rescale will be burned into pixel data!
[DICOM] Detected frame with shift/scale pair which exceeds the value range. Casting all frames to Float.
[3]:
vmin, vmax = image.min()[0], image.max()[0]
interact(
mpr_plot,
image=fixed(image),
labels=fixed(labels[0]),
x=IntSlider(112, 0, 191),
y=IntSlider(104, 0, 191),
z=IntSlider(195, 0, 226),
vmin=IntSlider(4000, vmin, vmax, 100),
vmax=IntSlider(36000, vmin, vmax, 100),
label_alpha=FloatSlider(0.7, min=0.0, max=1.0, step=0.1),
continuous_update=False
)
None
The ImageDescriptor describes the basic properties of the images. Each SharedImage also contains the modality information of its content.
[4]:
print(image.descriptor, '\n')
print(image.modality)
print(labels[0].modality)
imfusion.ImageDescriptor(dimensions: [w:192, h:192, s:227, c:1] | spacing:[3.64583, 3.64583, 3.27] | isMetric:true | type:FLOAT | memory: 31.92 MB)
Modality.NM
Modality.LABEL
Accessing image data with numpy
The imfusion-sdk package provides interoperability with numpy arrays. SharedImage and SharedImageSet can be converted to numpy arrays and the other way around.
The shape of a SharedImage is: [slices, width, height, channels].
[5]:
pet_image = pet.clone()[0]
array = np.array(pet_image)
print(pet_image.shape)
# select one slice for plotting
plt.imshow(array[::-1, 95, :, :], cmap='gray', vmin=0, vmax=15000)
plt.show()
(227, 192, 192, 1)
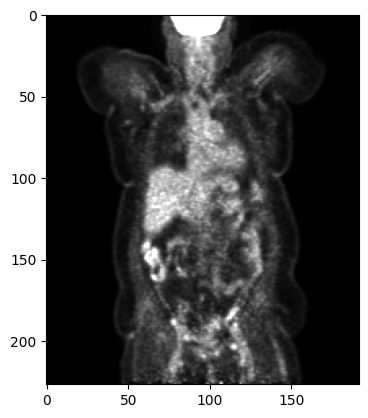
Since a SharedImageSet is basically a list of SharedImage, it has an additional dimension and the array shape is: [frames, slices, height, width, channels]
[6]:
array_set = np.array(pet, copy=False)
print(array_set.shape)
(1, 227, 192, 192, 1)
Modifying image data
np.array() creates a copy by default. Every change to the array is therefore not propagated to the SharedImage.
To update the SharedImage we have to copy the changed data back into the SharedImage using
assign_array().
Note: You cannot resize or reshape the image in this way, nor change the type of the image. In such cases you need to create a new image from the modified numpy array.
[8]:
# Create mask by thresholding
pet_image = pet.clone()[0]
array = np.array(pet_image, copy=False)
threshold = 10000
array[array < threshold] = 0
array[array >= threshold] = 1000
plt.imshow(array[::-1, 95, :, :], cmap='gray')
plt.show()
pet_image.assign_array(array)
mpr_plot(pet_image)
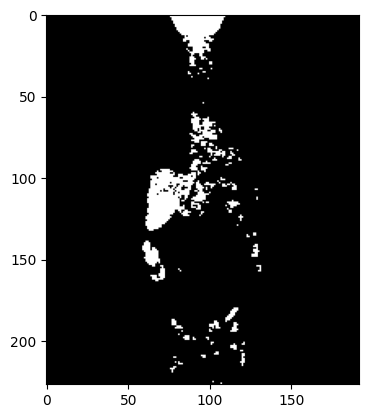

Save Images
A list of SharedImageSets can be saved to disk again …and then visualized in the ImFusion Suite
[9]:
# The generic `imfusion.save` method saves file to ImFusion files, which end with `.imf`
imfusion.save([pet], 'data/thresholded_pet.imf')
DICOM IO
The imfusion-sdk can also load DICOM files and folders and save specifically to DICOM format via a dedicated submodule called imfusion.dicom The module offers some extra IO routines specific to DICOM. For example IO from/to DICOM RTStructuredSet and DICOM Seg.
DICOM-RTStructure file IO
The following code shows how to load a DICOM RTStructure file and convert it to a labelmap (or binary mask).
[10]:
rtstructs = imfusion.load('data/pet-ct-rtstruct/uterus-rtstruct/1-1.dcm')
# rtstructs are represented as PointClouds
print("rtstruct", rtstructs)
# Convert them to labelmaps, by default each different set is converted to a separate mask
# Each mask has pixel value specified by the rtstruct label tag and some background
mask, *_ = imfusion.dicom.rtstruct_to_labelmap(rtstructs, pet)
print("mask", mask)
# Access the LabelDataComponent to get more information about the specific mask
ldc = mask.components.label
print(ldc.label_configs())
ldc = mask.components.label
print(ldc)
ldc.remove_label(0) # we don't care about the background here
print(ldc)
label_name = list(ldc.label_configs().values())[0].name
print(label_name)
tumor_mask = mask
assert tumor_mask is not None
rtstruct [<imfusion._bindings.PointCloud object at 0x7fdf65d352f0>]
mask imfusion.SharedImageSet(size: 1, [imfusion.SharedImage(UBYTE width: 192 height: 192 slices: 227 spacing: 3.64583x3.64583x3.27 mm)])
{0: LabelConfig(name="Background", color=(0.00, 0.00, 0.00, 1.00)), 1: LabelConfig(name="UTERUS - 1", color=(1.00, 1.00, 0.00, 1.00))}
<LabelDataComponent
0: LabelConfig(name="Background", color=(0.00, 0.00, 0.00, 1.00))
1: LabelConfig(name="UTERUS - 1", color=(1.00, 1.00, 0.00, 1.00))
>
<LabelDataComponent 1: LabelConfig(name="UTERUS - 1", color=(1.00, 1.00, 0.00, 1.00))>
UTERUS - 1
[11]:
mpr_plot(pet[0], labels=tumor_mask[0], y=100, z=195)
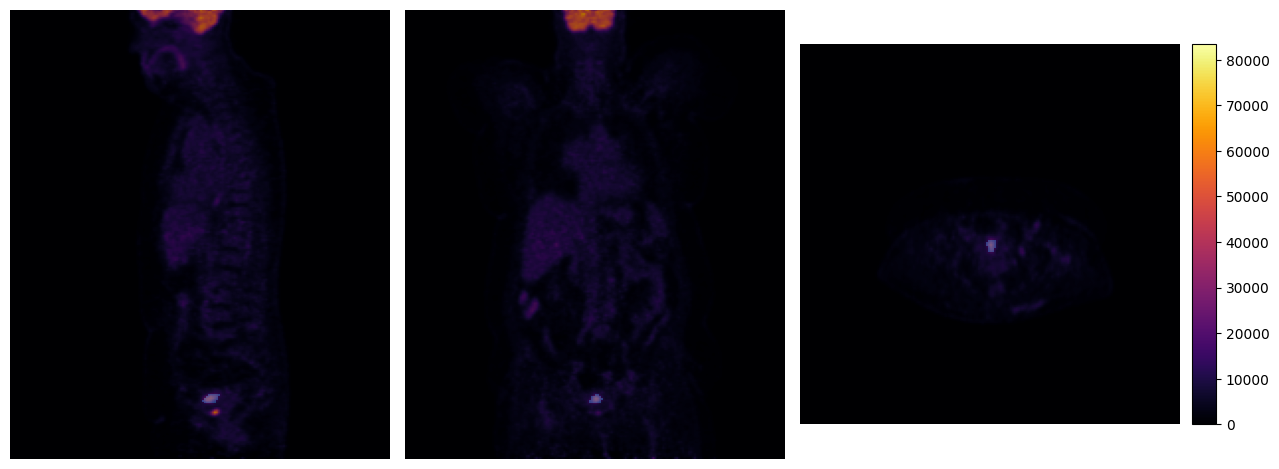
Saving a mask as RTStruct
[12]:
# Here we apply a dilation just to obtain a slightly different mask than the one we loaded
dilated_mask = ml.MorphologicalFilterOperation(mode='dilation', op_size=1, use_l1_distance=True).process(
tumor_mask) # dilate by 1 voxel
# save to rtstruct
imfusion.dicom.save_rtstruct(dilated_mask, pet, 'data/tumor_mask_dilated_rtstruct.dcm')
Saving a mask as DICOM Seg
[13]:
imfusion.dicom.save_file(tumor_mask, 'data/tumor_mask_seg.dcm', referenced_image=pet)
# Now load it again
(tumor_mask_from_dcm_seg,) = imfusion.dicom.load_file('data/tumor_mask_seg.dcm')
# Check that we have the same orientation
assert np.allclose(tumor_mask_from_dcm_seg[0].image_to_world_matrix, tumor_mask[0].image_to_world_matrix)
# assert the data is now bit exactly the original tumor mask
assert np.allclose(tumor_mask_from_dcm_seg[0].numpy(), tumor_mask[0].numpy())
[ ]: